Introduction #
TBX11K: Tuberculosis X-ray dataset contains 11200 X-ray images with corresponding bounding box annotations for tuberculosis (TB) areas, while the existing largest public TB datasets have much fewer X-ray images with corresponding image-level annotations. All images are with a size of 512x512. There are five main categories in this dataset: healthy, sick_but_non-tb, active_tb, latent_tb, and uncertain_tb. Also, the dataset includes the active_tb&latent_tb categorie. The authors split this dataset into train, val, and test sets, consisting of 6600, 1800, and 2800(3302) X-ray images, respectively. The proposed dataset enables the training of sophisticated detectors for high-quality computer-aided tuberculosis diagnosis (CTD). Authors reform the existing object detectors to adapt them to simultaneous image classification and TB area detection. These reformed detectors are trained and evaluated on the proposed TBX11K dataset and serve as the baselines for future research.
As a serious infectious disease, tuberculosis is one of the major threats to human health worldwide, leading to millions of deaths every year. Although early diagnosis and treatment can greatly improve the chances of survival, it remains a major challenge, especially in developing countries. Computer-aided tuberculosis diagnosis is a promising choice for TB diagnosis due to the great success of deep learning.
| Class | Train | Val | Test | Total | |
|---|---|---|---|---|---|
| Non-TB | Non-TB Healthy | 3000 | 800 | 1200 | 5000 |
| Sick & Non-TB | 3000 | 800 | 1200 | 5000 | |
| TB | Active TB | 473 | 157 | 294 | 924 |
| Latent TB | 104 | 36 | 72 | 212 | |
| Active & Latent TB | 23 | 7 | 24 | 54 | |
| Uncertain TB | 0 | 0 | 10 | 10 | |
| Total | 6600 | 1800 | 2800 | 11200 |
active_tb&latent_tb refers to X-rays that contain active and latent TB simultaneously. active_tb and latent_tb refer to X-rays that only contain active TB or latent TB, respectively.
Labels for active_tb:
- ActiveTuberculosis
latent_tb: - ObsoletePulmonaryTuberculosis
uncertain_tb refers to TB X-rays whose TB types cannot be recognized under today’s medical conditions. Uncertain TB X-rays are all put into the test set.
This is the distribution of the areas of TB bounding boxes. The left and right values of each bin define its corresponding area range, and the height of each bin denotes the number of TB bounding boxes with an area within this range. Note that X-rays are in the resolution of about 3000×3000. However, the original 3000×3000 images will lead to a storage size of over 100GB, which is too large to deliver. On the other hand, the authors found that the resolution of 512 × 512 is enough to train deep models for TB detection and classification. In addition, it is almost impossible to directly use the 3000 × 3000 X-ray images for TB detection due to the limited receptive fields of the existing CNNs. Therefore, the authors decided to only release the X-rays with a resolution of 512×512. For a fair comparison, they recommend all researchers use this resolution for their experiments.
Here are the answers from the dataset’s authors to questions about how the GT labels were created:
- Are latent TB cases biologically confirmed? (by IFNg testing or tuberculin skin testing)
Yes, they are. Both active and latent TB cases are biologically confirmed
using the hospitals’ accurate clinical diagnosis technology, of course,
in the image level.
- If the cases are biologically positive for active TB, but does has CXRs regions suspicious for latent TB only, are they labeled as latent TB or active TB?
As clarified in section 3.1.3, the annotation is conducted under a
double-check rule: “Specifically, each TB X-ray is first labeled by a
radiologist who has 5-10 years of experience in TB diagnosis. Then, his
box annotations are further checked by another radiologist who has >10
years of experience in TB diagnosis. They not only label bounding boxes
for TB areas but also recognize the TB type (active or latent TB) for
each box. The labeled TB types are double-checked to make sure that they
are consistent with the image-level labels produced by the golden
standard. If a mismatch happens, this X-ray will be put into the
unlabeled data for re-annotation, and the annotators do not know which
X-ray was labeled wrong before. If an X-ray is incorrectly labeled twice,
we will tell the annotators the gold standard of this X-ray and ask them
to discuss how to re-annotate it.” Therefore, the final TB type must be
consistent with the golden standard.
- If the cases are biologically positive for active TB, but does not contain CXRs regions that are not suspicious for active TB, how are they labeled?
The answer is similar to that of the above (2). The image-level labels
are confirmed using the hospitals’ accurate clinical diagnosis technology
and thus reliable. In our annotation process, our experienced
radiologists did not happen to the situation that you said, after
discussion, the annotation still cannot be consistent with the gold
standard.
- How can the cases be both active TB and latent TB?
Of course, one TB region is either active TB or latent TB. However, note
that an X-ray would contain both active TB and latent TB regions. In our
dataset, each TB box only has one label of being active or latent, but a
TB X-ray would have both active TB and latent TB labels.
| Datasets | Year | Class | Label | Sample |
|---|---|---|---|---|
| MC | 2014 | 2 | Image-level | 138 |
| Shenzhen | 2014 | 2 | Image-level | 662 |
| DA | 2014 | 2 | Image-level | 156 |
| DB | 2014 | 2 | Image-level | 150 |
| TBX11K | 2020 | 4 | Bounding box | 11200 |
The proposed TBX11K dataset is much larger, better annotated, and more realistic than existing TB datasets, enabling the training of deep CNNs. First, unlike previous (Schenzen dataset and CXR digital image datasets) that only contain several tens/hundreds of X-ray images, TBX11K has 11,200 images that are about 17× larger than the existing largest dataset, i.e., Shenzhen dataset, so that TBX11K makes it possible to train very deep CNNs. Second, instead of only having image-level annotations as in previous datasets, TBX11K annotates TB areas using bounding boxes, so that future CTD methods can not only recognize the manifestations of TB but also detect the TB areas to help radiologists for the definitive diagnosis. Third, TBX11K includes four categories of healthy, active_tb, latent_tb, and sick_but_non-tb, rather than the binary classification for TB or not in previous datasets, so that future CTD systems can adapt to more complex real-world scenarios and provide people with more detailed disease analyses.
The ground truth of the test set will not be released, because it is an online competition for computer-aided tuberculosis diagnosis. To promote the development of this field, the authors suggest you use the train set for training and the val set for validation when tuning your model. When you submit your results of the testing set to the authors’ server, they suggest you train your model on the train+val set and test on the test set.
Summary #
TBX11K: Tuberculosis X-ray is a dataset for an object detection task. It is used in the medical research, and in the medical industry.
The dataset consists of 11702 images with 1211 labeled objects belonging to 2 different classes including ActiveTuberculosis and ObsoletePulmonaryTuberculosis.
Images in the TBX11K dataset have bounding box annotations. There are 10903 (93% of the total) unlabeled images (i.e. without annotations). There are 3 splits in the dataset: train (6600 images), test (3302 images), and val (1800 images). Alternatively, the dataset could be split into 6 categories: healthy (3800 images), sick_but_non-tb (3800 images), active_tb (630 images), latent_tb (139 images), active&latent_tb (30 images), and uncertain_tb (0 objects). The dataset was released in 2019 by the Nankai University and InferVision.
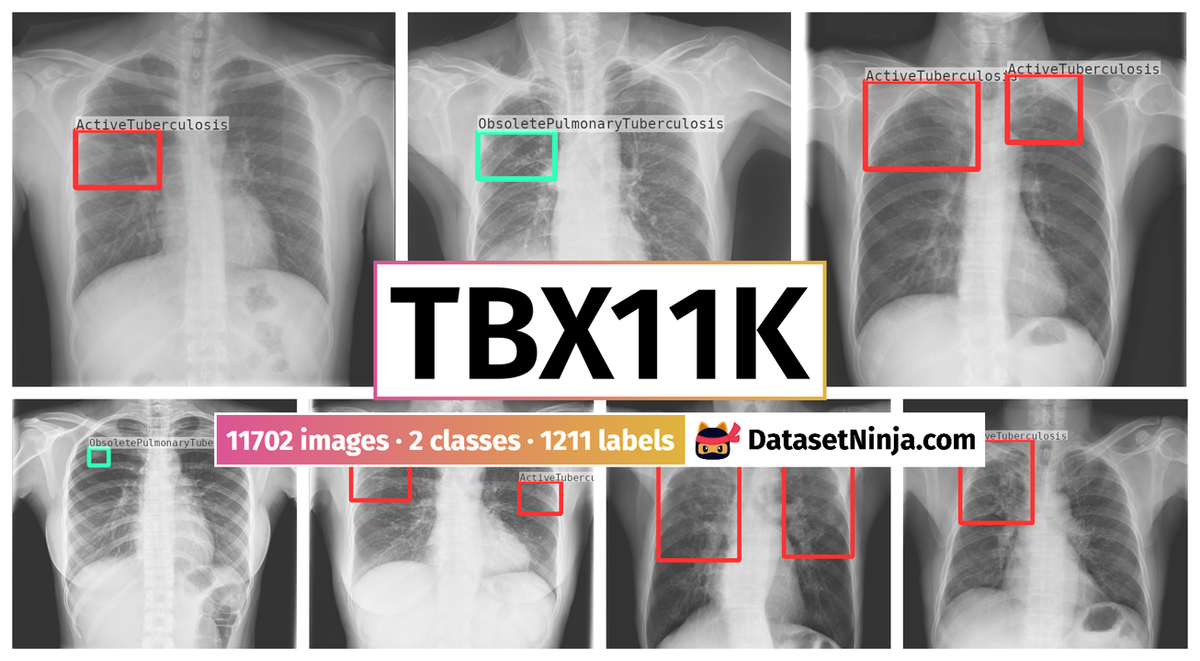
Explore #
TBX11K dataset has 11702 images. Click on one of the examples below or open "Explore" tool anytime you need to view dataset images with annotations. This tool has extended visualization capabilities like zoom, translation, objects table, custom filters and more. Hover the mouse over the images to hide or show annotations.
































Class balance #
There are 2 annotation classes in the dataset. Find the general statistics and balances for every class in the table below. Click any row to preview images that have labels of the selected class. Sort by column to find the most rare or prevalent classes.
Class ㅤ | Images ㅤ | Objects ㅤ | Count on image average | Area on image average |
|---|---|---|---|---|
ActiveTuberculosis➔ rectangle | 660 | 972 | 1.47 | 9.74% |
ObsoletePulmonaryTuberculosis➔ rectangle | 169 | 239 | 1.41 | 7.17% |
Co-occurrence matrix #
Co-occurrence matrix is an extremely valuable tool that shows you the images for every pair of classes: how many images have objects of both classes at the same time. If you click any cell, you will see those images. We added the tooltip with an explanation for every cell for your convenience, just hover the mouse over a cell to preview the description.
Images #
Explore every single image in the dataset with respect to the number of annotations of each class it has. Click a row to preview selected image. Sort by any column to find anomalies and edge cases. Use horizontal scroll if the table has many columns for a large number of classes in the dataset.
Object distribution #
Interactive heatmap chart for every class with object distribution shows how many images are in the dataset with a certain number of objects of a specific class. Users can click cell and see the list of all corresponding images.
Class sizes #
The table below gives various size properties of objects for every class. Click a row to see the image with annotations of the selected class. Sort columns to find classes with the smallest or largest objects or understand the size differences between classes.
Class | Object count | Avg area | Max area | Min area | Min height | Min height | Max height | Max height | Avg height | Avg height | Min width | Min width | Max width | Max width |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
ActiveTuberculosis rectangle | 972 | 6.62% | 26.66% | 0.23% | 23px | 4.49% | 364px | 71.09% | 132px | 25.81% | 26px | 5.08% | 212px | 41.41% |
ObsoletePulmonaryTuberculosis rectangle | 239 | 5.07% | 19.51% | 0.28% | 24px | 4.69% | 303px | 59.18% | 114px | 22.22% | 23px | 4.49% | 184px | 35.94% |
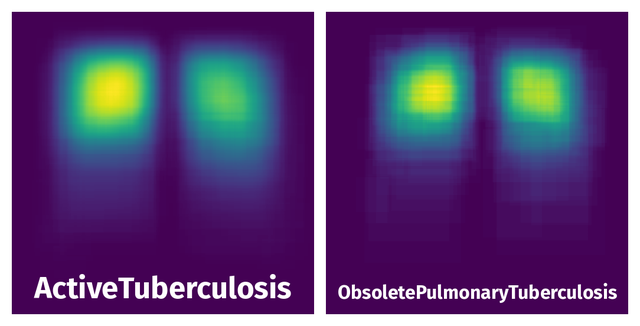
Spatial Heatmap #
The heatmaps below give the spatial distributions of all objects for every class. These visualizations provide insights into the most probable and rare object locations on the image. It helps analyze objects' placements in a dataset.
Objects #
Table contains all 1211 objects. Click a row to preview an image with annotations, and use search or pagination to navigate. Sort columns to find outliers in the dataset.
Object ID ㅤ | Class ㅤ | Image name click row to open | Image size height x width | Height ㅤ | Height ㅤ | Width ㅤ | Width ㅤ | Area ㅤ |
|---|---|---|---|---|---|---|---|---|
1➔ | ActiveTuberculosis rectangle | tb1042.png | 512 x 512 | 63px | 12.3% | 64px | 12.5% | 1.54% |
2➔ | ActiveTuberculosis rectangle | tb0680.png | 512 x 512 | 150px | 29.3% | 145px | 28.32% | 8.3% |
3➔ | ActiveTuberculosis rectangle | tb0680.png | 512 x 512 | 131px | 25.59% | 123px | 24.02% | 6.15% |
4➔ | ActiveTuberculosis rectangle | tb1150.png | 512 x 512 | 83px | 16.21% | 105px | 20.51% | 3.32% |
5➔ | ActiveTuberculosis rectangle | tb1150.png | 512 x 512 | 86px | 16.8% | 101px | 19.73% | 3.31% |
6➔ | ActiveTuberculosis rectangle | tb1150.png | 512 x 512 | 64px | 12.5% | 71px | 13.87% | 1.73% |
7➔ | ActiveTuberculosis rectangle | tb1150.png | 512 x 512 | 53px | 10.35% | 54px | 10.55% | 1.09% |
8➔ | ActiveTuberculosis rectangle | tb1168.png | 512 x 512 | 128px | 25% | 163px | 31.84% | 7.96% |
9➔ | ActiveTuberculosis rectangle | tb1028.png | 512 x 512 | 76px | 14.84% | 77px | 15.04% | 2.23% |
10➔ | ActiveTuberculosis rectangle | tb0564.png | 512 x 512 | 98px | 19.14% | 117px | 22.85% | 4.37% |
License #
Citation #
If you make use of the TBX11K data, please cite the following reference:
@inproceedings{liu2020rethinking,
title={Rethinking computer-aided tuberculosis diagnosis},
author={Liu, Yun and Wu, Yu-Huan and Ban, Yunfeng and Wang, Huifang and Cheng, Ming-Ming},
booktitle={Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition},
pages={2646--2655},
year={2020}
}
If you are happy with Dataset Ninja and use provided visualizations and tools in your work, please cite us:
@misc{ visualization-tools-for-tbx-11k-dataset,
title = { Visualization Tools for TBX11K Dataset },
type = { Computer Vision Tools },
author = { Dataset Ninja },
howpublished = { \url{ https://datasetninja.com/tbx-11k } },
url = { https://datasetninja.com/tbx-11k },
journal = { Dataset Ninja },
publisher = { Dataset Ninja },
year = { 2026 },
month = { feb },
note = { visited on 2026-02-28 },
}Download #
Dataset TBX11K can be downloaded in Supervisely format:
As an alternative, it can be downloaded with dataset-tools package:
pip install --upgrade dataset-tools
… using following python code:
import dataset_tools as dtools
dtools.download(dataset='TBX11K', dst_dir='~/dataset-ninja/')
Make sure not to overlook the python code example available on the Supervisely Developer Portal. It will give you a clear idea of how to effortlessly work with the downloaded dataset.
Disclaimer #
Our gal from the legal dep told us we need to post this:
Dataset Ninja provides visualizations and statistics for some datasets that can be found online and can be downloaded by general audience. Dataset Ninja is not a dataset hosting platform and can only be used for informational purposes. The platform does not claim any rights for the original content, including images, videos, annotations and descriptions. Joint publishing is prohibited.
You take full responsibility when you use datasets presented at Dataset Ninja, as well as other information, including visualizations and statistics we provide. You are in charge of compliance with any dataset license and all other permissions. You are required to navigate datasets homepage and make sure that you can use it. In case of any questions, get in touch with us at hello@datasetninja.com.


